A Molecular Network for Cognitive Decline in the Human Prefrontal Cortex
What's the science?
There are currently no therapies available to treat or prevent Alzheimer’s disease. This may be due to the complexity and heterogeneity of the disease. Although we know that the accumulation of beta-amyloid peptides and tau proteins occurs in the brain in Alzheimer’s disease and that certain susceptibility genes are involved, we do not understand the sequence of events that lead from genetic risk to brain pathology and cognitive decline. Brain gene expression data and network based analysis can potentially gather nuanced information about the complex interactions among genes that lead to brain pathology. This week in Nature Neuroscience, Mostafavi and colleagues use network-based analysis of gene expression data from the aging prefrontal cortex to elucidate a network involved in aging and Alzheimer’s disease.
How did they do it?
Prefrontal cortex RNA-sequencing (RNA-seq) data (gene transcript levels representing gene expression) from participants in longitudinal datasets of aging called the Religious Orders Study (ROS) and the Memory and Aging Project (MAP) were used. These datasets also contain post-mortem brain pathology data and longitudinal measures of cognitive performance. They ran a standard association analysis to look for genes whose expression levels associate with Alzheimer’s disease and cognitive decline. They then used an approach called gene module-trait network analysis that links key networks of genes that are co-expressed (related expression patterns) to cognitive decline and Alzheimer’s disease traits, and then selects the most strongly and directly associated networks. The goal of this was to gain more information on gene networks and their associated biological pathways than can be obtained from analyzing single gene-disease associations.
What did they find?
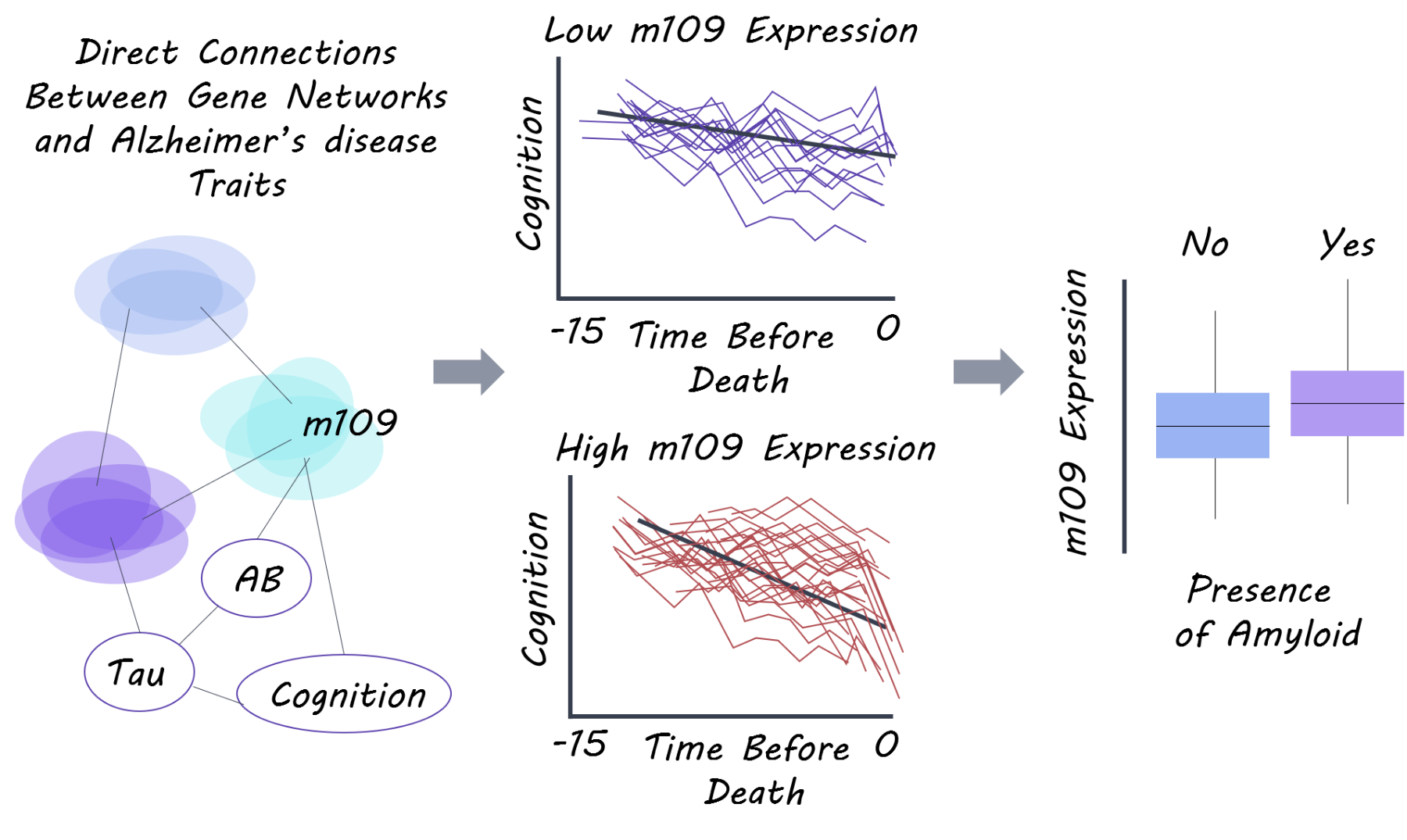
The known risk variant for Alzheimer’s disease in the APOE gene (strongest Alzheimer's risk gene) only explained 2.2% of the variance in Alzheimer’s disease (heterogeneity) and 5.1% of cognitive decline. When examining 21 other known Alzheimer’s risk variants, they only explain 2.1% of disease variance and 7.6% of cognitive decline, emphasizing that these genes alone do not explain disease heterogeneity and decline. In the gene module-trait network analysis, 47 modules of genes (from the RNA-seq gene expression data) were identified representing related networks of genes. 11 of these modules were associated with cognitive decline or Alzheimer’s disease related traits (beta-amyloid or tau), and they found that these modules replicated in their association (with Alzheimer’s disease) in an independent gene expression dataset.
They then used Bayesian network inference to determine direct gene module-trait associations while accounting for the heterogeneity of cell types in the brain (neurons, astrocytes etc). One module (module number 109) consisting of 390 genes involved in regulation of cell cycle and chromatin modification was most strongly associated with cognitive decline. They found that this module was strongly associated with beta-amyloid pathology. They then selected key genes from this module that were strongly expressed in neurons and astrocytes and showed the strongest gene-disease associations. They performed a knockdown experiment on these genes in astrocytes and stem cell derived neurons and found that knocking down 2 of these genes, INPPL1 (involved in lipid signalling) and PLXNB1 (involved in synaptic plasticity) significantly lowered beta-amyloid levels. INPPL1 explained 5.5% of variance in cognitive decline, while PLXNB1 explained 4.4%. They confirmed that these two genes drive a significant proportion of the effect of the m109 module (which explained 8.5% of cognitive decline) on beta-amyloid load, indicating that these two genes may be important for amyloid pathology and cognitive decline.
What's the impact?
This is the first study to use a network-based approach to identify direct gene network associations with Alzheimer’s disease traits and cognitive decline. This study identified a network of genes strongly associated with beta-amyloid load and cognitive decline, which are important measures of disease progression. This study demonstrates that a network-based approach can provide more information on networks of genes associated with cognitive decline that single gene associations might miss.
Mostafavi et al., A molecular network of the aging human brain provides insights into the pathology and cognitive decline of Alzheimer’s disease. Nature Neuroscience (2018). Access the original scientific publication here.